Medical Image Segmentation with Stochastic Aggregated Loss in a Unified U-Net
Published in 2019 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), 2018
Recommended citation: P. X. Nguyen, Z. Lu, W. Huang, S. Huang, A. Katsuki & Z. Lin (2019).Medical image segmentation with stochastic aggregated loss in a unified U-Net. In 2019 IEEE EMBS International Conference on Biomedical Health Informatics (BHI) (IEEE BHI 2019), Chicago, USA.
Paper Link: https://ieeexplore.ieee.org/document/8834667
Abstract
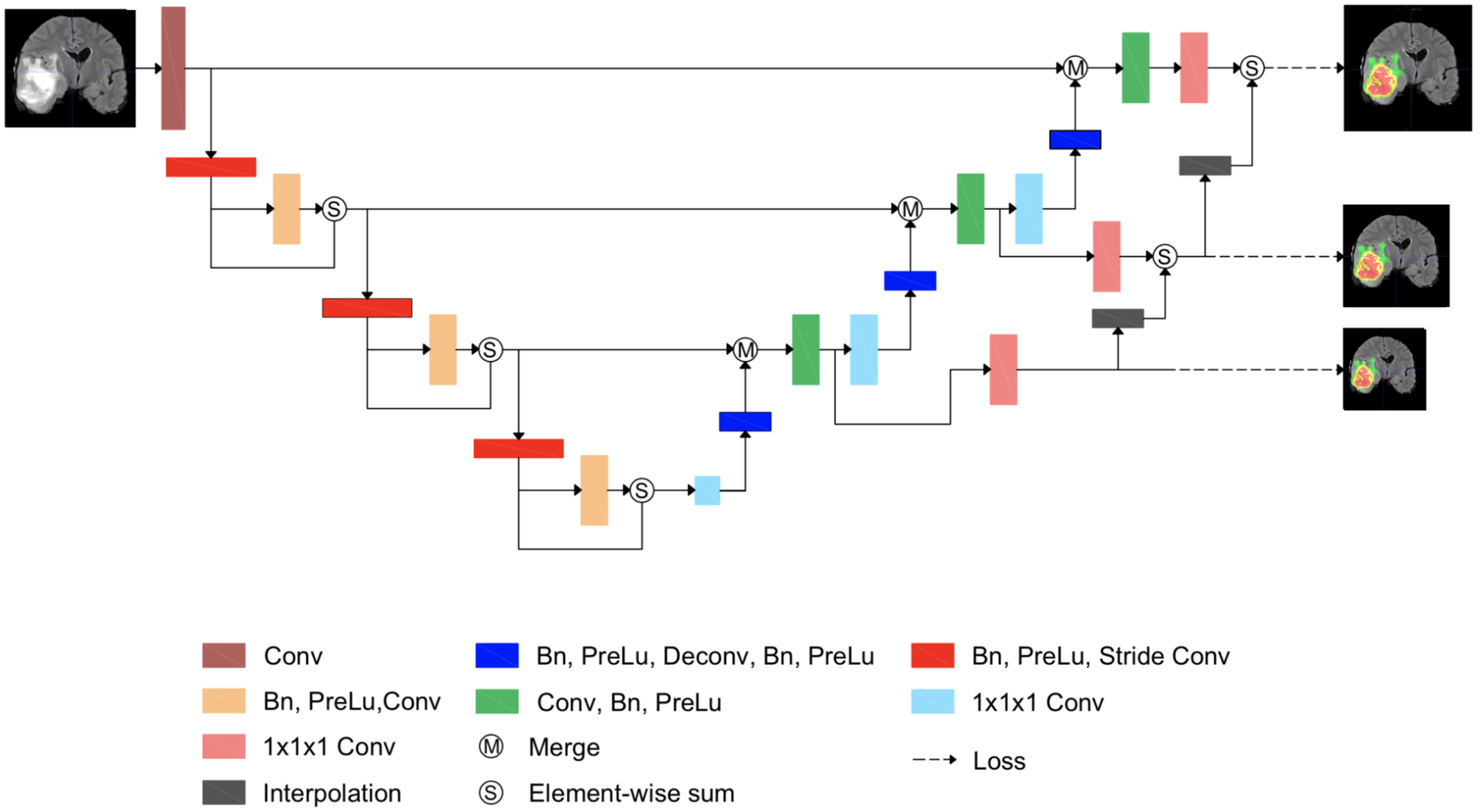
Automatic segmentation of medical images, such as computed tomography (CT) or magnetic resonance imaging (MRI), plays an essential role in efficient clinical diagnosis. While deep learning have gained popularity in academia and industry, more works have to be done to improve the performance for clinical practice. U-Net architecture, along with Dice coefficient optimization, has shown its effectiveness in medical image segmentation. Although it is an efficient measurement of the difference between the ground truth and the network's output, the Dice loss struggles to train with samples that do not contain targeted objects. While the situation is unusual in standard datasets, it is commonly seen in clinical data, where many training data available without the anomalies shown in the images, such as lesions and anatomic structures in some CTs/regions. In this paper, we propose a novel loss function - Stochastic Aggregated Dice Coefficient (SA Dice) and a modification of the network structure to improve its performance. Experimentally, in our own heart aorta CT dataset, our models beats the baseline by 4% in cross-validation Dice scores. In BRATS 2017 brain tumor segmentation challenge, the models also perform better than the state-of-the-art by approximately 2%.
Summary